Frequently Asked Questions
ModelMatcher is a global registry and search tool that is designed to facilitate collaborations between scientists and other stake holders of rare and undiagnosed disease research. While the primary goal of this project is to stimulate interactions between clinicians and basic scientists who are interested in the same/homologous genes, ModelMatcher can be used by patients, family members, funding agencies and industry. Our goal is to bring these communities together to facilitate mutually beneficial collaborative projects that advance scientific knowledge and patient care.
ModelMatcher is being developed by a group of scientists, clinicians and bioinformaticians who work at the Jan & Dan Duncan Neurological Research Institute at Texas Children’s Hospital and at Baylor College of Medicine. This team at the Texas Medical Center are working closely with the team developing the Rare Diseases Models & Mechanisms Network(RDMM) software at University of British Columbia. The Rare Diseases: Models and Mechanics software is open source and available at GitHub. You can learn more about the individual members on the “About Us” link at the top of the page.
Scientists register their human or model organism genes of interest in the "Scientist Registry". Users who wish to connect with these Scientists register their human genes of interest in the "Match Dashboard". ModelMatcher then generates a table in the Match Dashboard showing all the Scientists who share a gene of interest with them. The users are able to send official Match Requests to each Scientists via the Match Dashboard. The Match Dashboard will also keep track of when the last message was sent. The Scientist can then respond to the Match Request by contacting the Match Dashboard user through email.
Both the Scientist Registry and Match Dashboard pages of ModelMatcher are password-protected and the information you enter will be kept secure. The visibility of your information depends on your Data Privacy setting for each gene you select. The management team of the ModelMatcher project will have access to all data for administrative purposes.
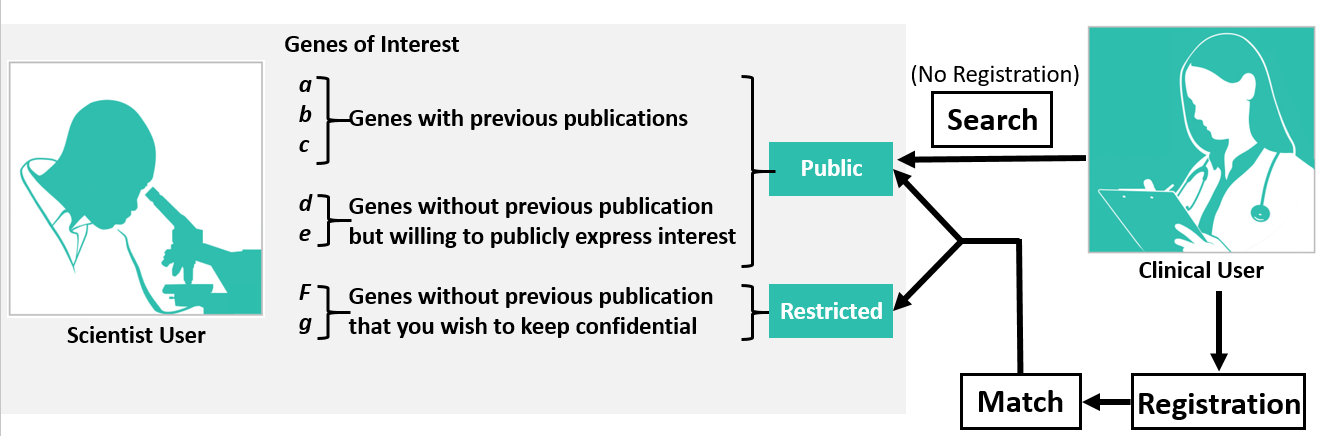
The profile information a Scientist enters in the Scientist Registry is made public, and can be accessed through both the "Search" and "Match" feature of ModelMatcher and partner registries. The visibility of your genes of interest can be optimized using your Data Privacy setting for each gene you select.
- Public - Your information will be visible publicly. Anyone can obtain information on these genes through ModelMatcher’s public search webpage. Your information will be shared (see above) with registered users and will be accessible to non-registered users through the registry’s public search webpage. This information will also be shared with our partner model organism registries including RDMM (Canada), Solve-RD (Europe), and AFGN (Australia) and our partners on the Matchmaker Exchange including GeneMatcher, PhenomeCentral and MyGene2.
- Restricted - A subset of genes of expertise/interest that can only be discovered by ModelMatcher users using the Match Dashboard feature, which requires the user to register with ModelMatcher or in one of our partner registries. . These typically include genes in which the Scientist user has unpublished data on and considers confidential.
The information users enters in the Match Dashboard will only be shared with Scientists when the user sends out match requests.
ModelMatcher is connected with model organism researcher registries around the world in order to maximize collaboration efforts. We share data between these registries so people on those systems can find collaborators on our system and vice versa. Currently, we partner with RDMM (Canada), Solve-RD (Europe) and AFGN (Australia), and this list may grow as more and more registries are established with similar goals and virtues.
ModelMatcher is connected to clinical registries that are part of the Matchmaker Exchange databases including GeneMatcher, PhenomeCentral and MyGene2 to bridge the basic scientific and clinical communities. If your registry wishes to connect with ModelMatcher, please contact us.
ModelMatcher currently supports research conducted on Human, Mouse, Rat, Frog, Zebrafish, Fruit Fly, Nematode Worm, Fission Yeast, Budding Yeast and E. coli genes. Even if a model organism of interest is not supported, Scientists can enter their gene of interest based on the Human homolog and this information will be indicated in the Scientist's Profile page.
You can add any gene that you are interested in establishing a collaboration for. If your lab actively works on a specific gene, you should list it as Primary (Tier 1) when you add it to your profile. The genes you are interested in but don't actively work on now can be listed as Secondary (Tier 2) genes. We recommend Scientists to add most genes as Public so you can be discovered through the Search feature of ModelMatcher, but you can also add genes as Restricted if you wish to identify collaborators on confidential projects.
- Public - Your information will be visible publicly. Anyone can obtain information on these genes through ModelMatcher’s public search webpage. Your information will be shared (see above) with registered users and will be accessible to non-registered users through the registry’s public search webpage. This information will also be shared with our partner model organism registries including RDMM (Canada), Solve-RD (Europe), and AFGN (Australia) and our partners on the Matchmaker Exchange including GeneMatcher, PhenomeCentral and MyGene2.
- Restricted - Genes that are classified as Restricted will only be used to perform matches with registered Match Dashboard users and users in our partner registries. If Scientist Users do not wish to publicly disclose that they are interested in certain genes (e.g. unpublished genes with preliminary data, genes in which collaboration is underway with confidentiality), we recommend they classify these genes into the Restricted category.

ModelMatcher has two tiers of gene expertise: those you currently work on, or those you could rapidly position yourself to work on. These are called Primary (Tier 1) or Secondary (Tier 2).
Primary genes (tier 1 genes, genes you currently work on): These are genes which your lab actively works on, and you have most reagents, assays and protocols ready to perform functional assays
Secondary genes (tier 2 genes, genes you currently don't work on): These are genes which your lab has worked on in the past or have strong interest in, but would need additional time to establish protocols or create reagents to perform functional assays
No. When you are adding model organism genes, please use the official gene symbol for the gene you study in that model organism. The only time you would need to identify a human homolog and input that gene is if you work in a model organism not supported by ModelMatcher.
ModelMatcher uses DIOPT to find homologs to human genes. Currently, we use a score > 5 and best score in either forward or reverse as cutoffs for homologous genes. For some species that are not covered by DIOPT (e.g. E. coli) or there are true homologous gene pairs with low DIOPT scores, we try to manually add them to our list of potential ortholog candidates (the taxon and gene ID's in this file are all NCBI format). We are always trying to improve our system, so if you notice a homologous gene is not coming up, or a non-homologous gene is, please reach out to us using the feedback feature. Our team will review the relevant data and change our homolog outputs to increase our accuracy!
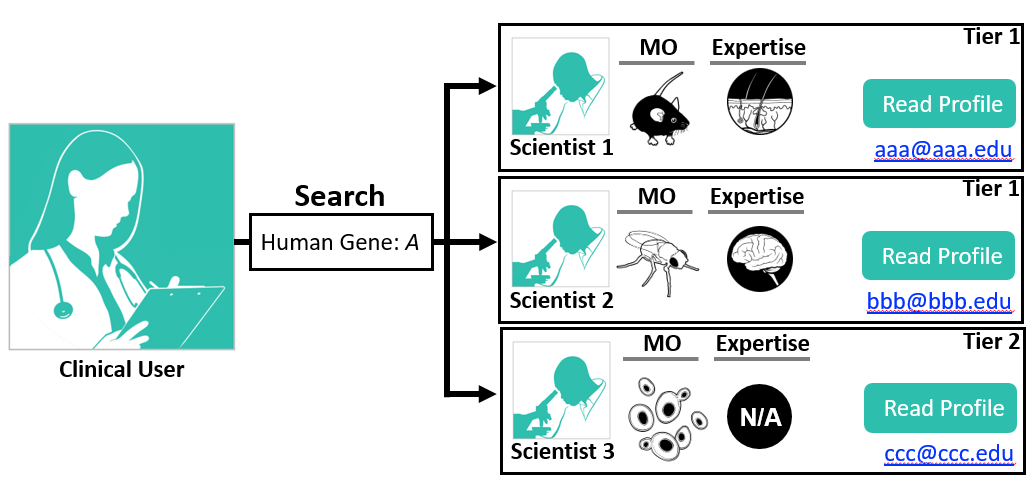
The Search page allows you to search Scientists who chose to publicly share their genes of interest. users can also connect to partner registries including RDMM (Canada), Solve-RD (Europe), and AFGN (Australia) and display/search their public information from their Scientists users.
After clicking on the Search button, the list of Scientists and their information summary will be displayed at the bottom of the page. Some scientists, who registered the gene as Restricted, are listed as anonymous. These scientists can be contacted through the Match Dashboard. Our partner registries results will be displayed in a separate section. Clicking on the "view user profile" button will open a new tab with the user's full profile and gene information.
Gene symbols are organism-specific and will be matched against the list of known gene symbols for the selected organism (selection made in a dropdown menu). Currently, we use the NCBI Gene symbol for all organisms. As a default, users can enter a human gene symbol and obtain results for all supported model organisms. To learn about how ModelMatcher identifies potential orthologs, please visit the "How does ModelMatcher determine model organism homologs?" FAQ entry.
The Match Dashboard is our platform designed for user who wish to connect with other Scientists, Clinicians and Patients. Users of this service will create a profile which includes their name, user type (clinician, patient, non-profit, etc.) and an email address. The user then inputs human genes they are interested in. Under the -Match with Scientist- tab, a table of predicted homologs and the Scientists who work on these genes is generated from data stored in the Scientist Registry. From this table, users can send official Match Requests to each Scientist. This is the only way to reach out to anonymous Scientists who have registered a specific gene under the Restricted privacy setting. If the scientist wishes to collaborate, they will respond to the email address associated with your Match Dashboard Account. In addition, the -Match with Clinicians and Patients- tab will allow users to find clinical collaborators through the Matchmaker Exchange.
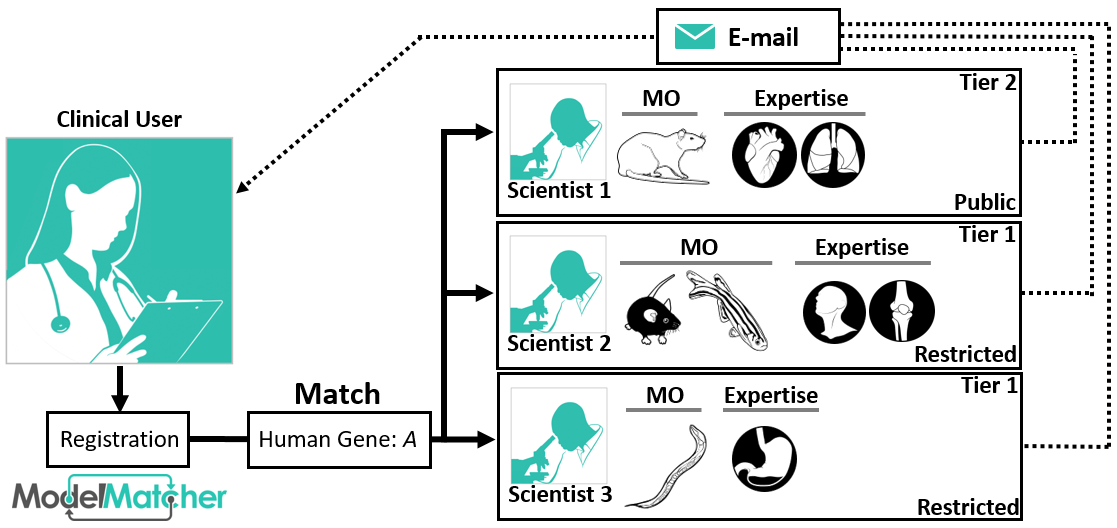
If you are using the public search function, you can attempt to contact them with the information listed on their profile. Registered users can initiate a Match Request from their Match Dashboard where they can write a short message about their interest in this gene. ModelMatcher will generate an official email detailing who you are, your interest in the gene, and ways to contact you. Users will receive a confirmation email when the message is sent to the Scientist, and the date of last contact is displayed on the Match Dashboard.
Yes, but there are some rules! Our Model Organism, Model System and Organ System icons are registered under Creative Commons BY-NC 4.0. This means you are free to use them in any non-commercial way as long as you credit ModelMatcher. Image files of each icon used on this website can be found here.
Anyone proposing to use data shared through MyGene2 in a publication agrees to:
- The user will contact the owner of the matching dataset to assess the integrity of the match and if validated will offer appropriate agreed recognition of their contribution, which may include co-authorship if the magnitude of the contribution warrants it.
- Acknowledge MyGene2 using the following wording: “This study makes use of data shared through MyGene2.org. Funding for MyGene2 was provided by the National Institutes of Health / National Human Genome Research Institute and National Heart Lung and Blood Institute through the University of Washington Center for Mendelian Genomics (2UM1HG006493-05) and by Wellcome Trust, U.S. National Institutes of Health, Howard Hughes Medical Institute through the Open Science Prize.”
MyGene2 provides data in good faith as a investigative tool, but without verifying the accuracy, clinical validity or utility of the data. MyGene2 makes no warranty, express or implied, nor assumes any legal liability or responsibility for any purpose for which the data are used.
The user certifies that no attempt to identify individual patients from other MatchMaker Exchange databases will be undertaken. The user certifies that no attempt to identify individuals in MyGene2 if the individuals have not chosen to identify themselves.
PhenomeCentral provides data in good faith as a research tool, but without verifying the accuracy, clinical validity or utility of the data. PhenomeCentral makes no warranty, express or implied, nor assumes any legal liability or responsibility for any purpose for which the data are used.
The user certifies that no attempt to identify individual patients will be undertaken.
Prior to using PhenomeCentral data in a publication, the user will contact the owner of the matching dataset to assess the integrity of the match and if validated will offer appropriate agreed recognition of their contribution, which may include co-authorship if the magnitude of the contribution warrants it to at least one representative from the project/participating centre (possibly the member who submitted the patient data).
If PhenomeCentral enables a research publication, the authors must acknowledge PhenomeCentral using the following wording: “This study makes use of data shared through the PhenomeCentral repository. Funding for PhenomeCentral was provided by Genome Canada and Canadian Institute of Health Research (CIHR).”
People proposing to publish material that makes use of matches achieved through the GeneMatcher database agree to: Acknowledge GeneMatcher and the grant that supports it: 1U54HG006542; and Notify the systems administrator of a published paper (so that we can track our results), by sending an email through the "Contact Us" tab on GeneMatcher. No one is authorized to attempt to identify patients whose results are in GeneMatcher by any means. This applies to matches made within GeneMatcher or with any other database included in the Matchmaker Exchange. GeneMatcher provides these data in good faith as a research tool, but without verifying the accuracy, clinical validity or utility of the data. GeneMatcher. Johns Hopkins University makes no warranty, express or implied, nor assumes any legal liability or responsibility for any purpose for which the data are used.When referring to ModelMatcher, please cite the following preprint article in BioRxiv.
ModelMatcher: A scientist-centric online platform to facilitate collaborations between stakeholders of rare and undiagnosed disease research
J. Michael Harnish, Lucian Li, Sanja Rogic, Guillaume Poirier-Morency, Seon-Young Kim, Undiagnosed Diseases Network, Kym M. Boycott, Michael F. Wangler, Hugo J. Bellen, Philip Hieter, Paul Pavlidis, Zhandong Liu, Shinya Yamamoto. bioRxiv 2021.09.30.462504; doi: https://doi.org/10.1101/2021.09.30.462504
Glossary
Scientists: Users who are scientists with expertise in specific genes who access ModelMatcher. Scientists who register in ModelMatcher are willing to use in vivo, in vitro, and/or in silico methods to experimentally study the function of genes and variants linked to human diseases.
Non-Scientist Users: Users with a clinical interest who access ModelMatcher to identify scientific collaborators. These users may be clinicians (e.g. physicians, genetic counselors) who are trying to diagnose a rare disease patient, patients themselves, or their family members who wish to participate in research to understand the mechanism of their disease. Funding agencies and non-profit organizations can use ModelMatcher as non-Scientist Users to identify a group of scientists with expertise in a specific gene to facilitate collaborative research.
Genes of expertise/interest: A list of genes that a Scientist User registers in ModelMatcher as having expertise or interest in.
Public genes: A subset of genes of expertise/interest that can be discovered by non-Scientist Users using the “Search” feature, which does not require the user to be logged in or registered with ModelMatcher. These typically include genes in which the Scientist User has published on the past and considers non-confidential.
Restricted genes: A subset of genes of expertise/interest that can only be discovered by Match Dashboard Users using the "Match Request" feature, which requires the user to register with ModelMatcher or other federated matchmaking databases. These typically include genes in which the Scientist User has unpublished data on and considers confidential.
Primary genes (tier 1 genes, genes Scientist Users currently work on): : These are genes which Scientist Users' labs actively work on, and have reagents and protocols ready to perform functional assays.
Secondary genes (tier 2 genes, genes Scientist Users currently don't work on): These are genes which Scientist Users' labs have worked on in the past or have strong interest in, but would need additional time to establish protocols or create reagents to perform functional assays.
Match Dashboard: A personalized page for users to register their genes of interest to identify potential collaborators in ModelMatcher and our partner registries. Users can reach out to scientists, clinicians, and patients by submitting "Match Requests". This is the only way a user can reach out to Scientists who registered their genes of interest as "Restricted".